Research ArticleOpen Access, Volume 2 Issue 1
Antimicrobial Susceptibility Profiles of Diarrheagenic Escherichia coli Strains Isolated in Children Below Five Years and Food Animals Kisumu County, Kenya
Redemptah Yeda1*; George Makalliwa1 ; Caroline Ouma2 ; Elekiah Anguko2 ; John Gachohi1,3,4; Gideon Kikuvi1
1School of Public Health, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya.
2Department of Diagnostic and Laboratory Systems Program, Center for Disease Control, Kisumu, Kenya.
3Washington State University Global Health Program, Washington State University, P.O. Box 72938, Nairobi 00200, Kenya.
4Paul G Allen School of Global Health, Washington State University, Pullman WA99164, USA.
*Corresponding author: Redemptah Yeda
School of Public Health, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya.
Received : Jan 04, 2024 Accepted : Feb 19, 2024 Published : Feb 26, 2024
Epidemiology & Public Health - www.jpublichealth.org
Copyright: Yeda R © All rights are reserved
Citation: Yeda R, Makalliwa G, Ouma C, Anguko E, Gachohi J, et al. Antimicrobial Susceptibility Profiles of Diarrheagenic Escherichia coli Strains Isolated in Children Below Five Years and Food Animals Kisumu County, Kenya. Epidemiol Public Health. 2024; 2(1): 1035.
Abstract
Background: Diarrheagenic Escherichia coli (DEC) are the major cause of diarrhoea in children in developing and undeveloped countries. Domestic food animals act as reservoirs of DEC that are transmitted to children below five years through inadequate hygiene practices.
Aim: To determine the antimicrobial susceptibility patterns of DEC strains isolated from children below five years and food animals in Kisumu County.
Methods: A total of 250 samples, were collected and analyzed using polymerase chain reaction. The antimicrobial susceptibility was determined using Kirby-Bauer disc diffusion method.
Results: Enteroaggregative E.coli (12%), Enteropathogenic E.coli (5%), Enterotoxigenic E.coli (3%) and mixed infections (3%) respectively. From the DEC children sample isolates, antimicrobial agents showed 84.6% sensitivity towards Meropenem followed by Gentamicin (80.8%) and then Nalidixic acid (73.1%). High resistance was recorded among the Tetracycline and Sulfamethoxazole antibiotics. Of the 26 DEC children sample isolates, 24(92%) exhibited multidrug resistance patterns: 46% showed resistance to four different antibiotics, 38% to five different antibiotics and 4% to six different antibiotics. Animal samples showed 100% susceptibility to most antibiotics, with only two isolates showing multidrug resistance to five antibiotics and one showing resistance to four and three antibiotics, respectively. The multidrug index value for the DEC pathotypes isolated in children was 0.5, indicating a high risk of DEC transmission.
Conclusion: Multidrug resistant bacteria reported in food animals and children populations suggested the possibility of transmission of multidrug resistance bacteria between children and food animals. Other sources of transmission also need to be investigated.
Keywords: Diarrheagenic Escherichia coli; Multidrug resistance; Multidrug index.
Background
The increasing number of bacterial infections represent the global health crisis regarding infectious diseases [1]. Animals are always proposed as pathogenic bacterial reservoirs and may play a critical role in transmission to humans [2]. The interaction between animals and humans allows both populations to act as reservoirs from which antimicrobial-resistant bacterial infections can be bidirectionally transmitted [3]. The role of animals in the emergence and transmission of antimicrobial resistance to the human population is yet to be fully understood.
Improper use of antimicrobial agents by humans are considered major drivers of antimicrobial resistance [4]. Escherichia coli (E.coli) is a bacteria that normally lives in the intestines of both healthy people and animals. The prevalence of antibiotic resistance among E. coli isolates from patients with acute diarrhoea in Egypt was 68.2%, 57.2%, and 24.2% for ampicillin, trimethoprim-sulfamethoxazole, and ampicillin respectively [5]. Studies conducted in Asia have reported the highest resistance to the penicillin class of antibiotic with amoxicillin recording 80.9% followed by ampicillin at 73.5% [6]. Escherichia Coli also represents a major reservoir of AMR genes that may be responsible for treatment failures in both human and animals. An increasing number of AMR genes has been identified in E. coli many of which are located in mobile genetic elements and hence can be transmitted between animals, humans and the natural world [7].
AMR represents a global public health problem, for which the patterns among bacteria isolated from children with close contact to food animals is not widely documented. Hence, this study a clear understanding of the antibiotic profiles of both children below five years and food animals in Kisumu County. The data generated from this study would establish epidemiological surveillance systems that would promote collaboration directed on the wellbeing of children below five years and animals
Materials and methods
Study area: This study was carried out in Kisumu County from August 2022 to February 2023. Kisumu has an annual rainfall of 276.22 mm and temperature of 23.93o C. It sits at an elevation of 1,131 m (3,711 ft). Kisumu is 200 miles northwest of Nairobi and it is located at the shores of lake Victoria.
Study population: All children below five years with diarrhea who attended Kisumu County hospital and had a food animal kept at their homes were included in the study
Study design: This was a cross sectional study conducted in Kisumu County Hospital and this was chosen because of availability of clinical cases (children with diarrhea) and willingness of the childrens’ parents to consent.
Sample size determination: Prevalence based methodology was used to estimate burden of diarrhea disease in Kisumu County among children below five years. Kisumu County reports diarrhea prevalence of 18% higher than the neighboring areas [8]. The P-value of less than 0.05 were considered statistically significant with a detection rate at 18% for children below 5 years with diarrhea [9].
The sample size was determined using the Fishers method.
Sample size N0=Z2pq/d2
Where: n = desired minimal sample size (when population is greater than 10,000)
z = standard normal deviate which is 1.96 at 95% confidence level.
p = estimated prevalence of diarrheal disease among under five children in Kisumu (18%).
d = degree of accuracy (0.05)
q = 1-p
Hence, using a confidence of 95% that corresponds to the standard normal deviate of 1.96, the proportion in the target population estimated at 18% and the degree of accuracy required set at 0.05, the sample size is:
=1.962*0.18*0.82
0.052
=227
10% of 227+23=250 (The 10% was to cover for lost samples during processing) = 250
Therefore a total of 250 samples were collected from both children and animals residing in Kisumu County, Kenya.
Sample collection: The sample collection procedure was done by a certified medical laboratory technician having the stool laboratory request from the pediatrician for the diarrheal child below five years who visited Kisumu County Hospital. The stool samples for children below two years who were unable to give stool at the moment were collected by researchers from the diapers, the stool samples were well labelled and aseptically transferred into a Cary Blair tube and transported to the Kenya Medical Research Laboratory in an ice box containing ice packs for bacterial isolation.
Isolation and identification of diarrheagenic E. coli: Isolation of E.coli was done on MacConkey agar, each of the 250 correctly collected fecal sample was streaked onto the MacConkey plate using a sterile wire loop in a biosafety cabinet, following incubation at 370 C for 18-24 hours. Pink colonies (lactose fermenters) and pale colonies (non-lactose fermenters) were observed. Three to five well separated lactose fermenting colonies were sub-cultured in a nutrient agar taken as pure E.coli culture. Each culture was then subjected to standard biochemical testing for identification of suspected E.coli. The pure E. coli isolates were suspended in tryptone soy broth supplemented with 20% glycerol and stored at -80°C awaiting subsequent analysis.
DNA isolation: The first four colonies of E.coli from overnight growth on MacConkey (Oxoid, Basingstoke, UK) were suspended in 200 µl of nuclease-free water and mixed thoroughly using a vortex. This was followed by boiling the bacterial suspension at 95o C for 20 minutes. The rich DNA supernatant was harvested after centrifugation at 12,000 ×g for 10 minutes and stored at -20°C[10].
Multiplex polymerase chain reaction: Characterization of the three selected E.coli pathotypes; EAEC, EPEC, and ETEC, was carried out using a multiplex PCR. The 25 μl PCR reaction mixture contained 12.5 μl of 2X DreamTaq Green PCR Master Mix (Thermo Scientific, Waltham, MA, USA), 0.5 μl (30 μM) each of forward and reverse primers, 6.5 μl of nuclease-free water and 5 μl of DNA template. The PCR reaction was performed on a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA, USA) with an initial denaturation of 95°C for 5 min, followed by 25 cycles of amplification (94°C for 1 min, 58°C for 1:30 min and 72°C for 1:30 min) and a final extension step at 72°C for 10. Positive and negative controls were ATCC 25922 strain and nuclease-free water, respectively.
Antimicrobial susceptibility testing: The antibiotic susceptibility profiles of confirmed Diarrheagenic strains were determined against eleven antibiotic (Table 1). Using the disk diffusion method on Muller Hinton Agar (Oxoid, Basingstoke, Hampshire England) as described by [11]. The E. coli ATCC 25922 was used for quality control. The diameters of the zones of inhibition were measured using a Vernier caliper and classified as sensitive, Intermediate and resistant according the clinical and laboratory standard institute guidelines (CLSI, 2022). Multidrug resistant Index (MDRI) of individuals isolates was calculated by dividing the number of antimicrobial agents to which the isolates were resistant by the total number of drugs to which isolates were tested against. Values lower than 0.2 were considered low risk while values higher than 0.2 were considered high risk [12].
Table 1: Antimicrobial discs used for susceptibility testing of diarrheagenic E. coli Isolates.
| Antibiotic class | Antimicrobial agent (Potency) |
|---|---|
| Aminoglycosides | Gentamycin (10 μg) |
| Carbapenems | Meropenem (10 μg) |
| Penicillin | Ampicillin (10 μg), Amoxillin (10 μg) |
| Beta lactam combination | Amoxicillin |
| Cephems | Ceftriaxone (30 μg) |
| Phenicols | Chloramphenicol (30 μg) |
| Sulphonamides | Trimethoprim- sulfamethoxazole (25 μg) |
| Quinolones | Nalidixic acid (30 μg) |
| Tetracycline | Tetracycline (30 μg) |
| Macrolides | Azithromycin (15 μg) |
| Fluoroquinolones | Ciprofloxacin (5 μg) |
Data analysis: All data from the laboratory tests were coded, filtered, and recorded in Microsoft Excel spreadsheet 2007 (Microsoft Corporation) before being analyzed with Stata version 15.1. All data from diarrheic children of under the age of five were subject to contingency table analysis, and Pearson’s Chisquare test was used to determine the statistical significance of the relationships and/or associations between the dependent and independent variables from the data. To examine the relationship between each predictor variable and the result variable, a logistic regression analysis was used. Throughout the investigation, a significance level of p<0.05 at 95% confidence intervals was employed.
Results
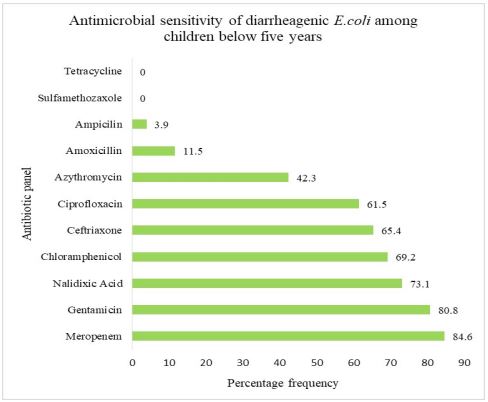
Antimicrobial sensitivity of diarrheagenic Escherichia coli isolated from children aged below five years: The antimicrobial sensitivity patterns of 26 diarrheagenic E. coli showed highest sensitivity patterns to various antibiotics; Of these 22(84.6%) diarrheagenic E. coli positive isolates were sensitive to Meropenem, 21(80.8%) of the diarrheagenic E. coli were sensitive to Gentamicin, 19(73.1%) diarrheagenic E. coli positive isolates showed sensitivity towards Nalidixic acid and 18(69.2%) of the diarrheagenic E. coli positive isolates were sensitive to Chloramphenicol Figure 2.
Table 2: Multiple drug-resistant DEC isolates among children.
| No of antibiotic-resistant | No of isolates (24) | Percentage |
|---|---|---|
| 3 | 3 | 12% |
| 4 | 11 | 46% |
| 5 | 9 | 38% |
| 6 | 1 | 4% |
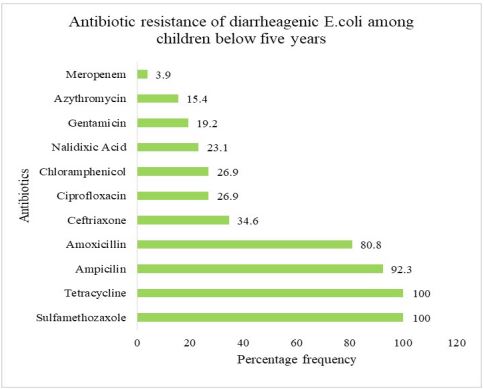
Antimicrobial resistance patterns of diarrheagenic Escherichia coli isolated from children aged below five years: The antimicrobial resistance of 26 diarrheagenic E. coli isolates showed 100% resistance to tetracycline and SulfamethoxazoleTrimethoprim. Of the positive diarrheagenic E. coli isolates, 24(92.3%) showed resistance towards ampicillin and 21(80.8%) showed resistance to Amoxicillin (Figure 3).
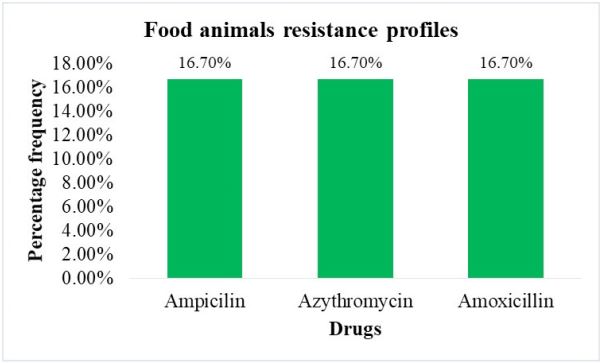
Antimicrobial resistance patterns of diarrheagenic Escherichia coli from food animals: Of the six food animal samples that were positive for various diarrheagenic E.coli strains 3/6 isolates showed resistance lower than 20% towards the three drugs Amoxicillin, Azithromycin and Ampicillin. They showed 100% sensitivity among the other eight antibiotics tested (Figure 4).
Multidrug resistance among DEC isolates from children: Of the 26 DEC isolates 24 showed multidrug resistance to more than three different drugs (Table 2).
Table 3: Antimicrobial resistance percentage of diarrheagenic Escherichia coli strains/virulence genes.
| Drugs | EAEC | EPEC | ETEC | MIXED INFECTIONS | P-value (95%CI) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistance | Intermediate | Sensitivity | Resistance | Intermediate | Sensitivity | Resistance | Intermediate | Sensitivity | Resistance | Intermediate | Sensitivity | ||
| Chloramphenicol | 21.4% | 0 | 78.6% | 33.3% | 33.3% | 33.3% | 20% | 0 | 80% | 50% | 0 | 50% | 0.134 |
| Sulfamethoxazole | 100% | 0 | 0 | 100% | 0 | 0 | 100% | 0 | 0 | 100% | 0 | 0 | - |
| Tetracycline | 100% | 0 | 0 | 100% | 0 | 0 | 100% | 0 | 0 | 100% | 0 | 0 | - |
| Ciprofloxacin | 14.3% | 14.3% | 71.4% | 66.7% | 33.3% | 0 | 20% | 0 | 80% | 50% | 0 | 50% | 0.219 |
| Nalidixic Acid | 21.4% | 0 | 78.6% | 33.3% | 0 | 66.7% | 40% | 20% | 40% | 0 | 0 | 100% | 0.31 |
| Ampicillin | 85.7% | 7.1% | 7.1% | 100% | 0 | 0 | 100% | 0 | 0 | 100% | 0 | 0 | 0.932 |
| Gentamicin | 7.1% | 0 | 92.9% | 33.3% | 0 | 66.7% | 20% | 0 | 80% | 50% | 0 | 50% | 0.247 |
| Azithromycin | 14.3% | 42.9% | 42.9% | 33.3% | 66.7% | 0 | 20% | 20% | 60% | 0 | 50% | 50% | 0.665 |
| Meropenem | 0 | 14.3% | 85.7% | 33.3% | 33.3% | 33.3% | 0 | 0 | 100% | 0 | 0 | 100% | 0.082 |
| Amoxicillin | 78.6% | 7.1% | 14.3% | 66.7% | 0 | 33.3% | 80% | 20% | 0 | 100% | 0 | 0 | 0.655 |
| Ceftriaxone | 21.4% | 0 | 78.6% | 66.7% | 0 | 33.3% | 40% | 0 | 60% | 50% | 0 | 50% | 0.404 |
Discussion
The present study has revealed sensitivity to Meropean, Gentamicin and Nalidixic acid above 70% among children below five years residing in Kisumu County. However there was resistance to Tetracycline, Sulfamethoxazole-trimethoprim and Amoxicillin above 90%. Isolates from animal samples showed sensitivity to most antibiotics and reported low levels of resistance below 20% towards Ampicillin, Amoxicillin and Azithromycin. Our analysis has shown that poor sanitation and environment contamination could link AMR transmission between animals and humans and could have contributed to increase in disease burden and demand of antibiotics among the children below five years.
All the Enteroaggregative E. coli strains in diarrheal children were resistant to tetracycline and Sulfamethoxazole –trimethoprim while 85.7% and 78.6% were resistant to Ampicillin and Amoxicillin respectively. The Enterotoxigenic and Enteropathogenic E. coli strains in diarrheal children showed (100%) resistance to Sulfamethoxazole-trimethoprim, tetracycline and ampicillin while resistance to Amoxicillin was 66.7% and 80% respectively. While the mixed infection (EAEC/ETEC and EAEC/ EPEC) samples showed high level of resistance to Sulfamethoxazole –trimethoprim, Tetracycline, Ampicillin and Amoxylin (100%) (Table 3).
The present study reports DEC pathotypes resistant to Tetracycline and Sulfamethoxazole (100% 26/26) and (92.3% 24/26), Ampicillin (80.8% 21/26), Amoxicillin (34.6% 9/26) and Ceftriaxone (26.9% 7/26). The resistance reported could be problematic in the management of bacteria-caused diarrhea. DEC pathotypes were also found resistant to these commonly prescribed drugs in other studies, including Ethiopia [13] and Nigeria [14]. Our findings also tend to concur with a study by Verma et al. in India that have reported high resistance to Trimethoprim 96% and tetracycline and ampicillin 97.3% [15]. This concurs with studies performed in Nairobi Kenya [16]. However, compared to other studies, prevalence to tetracycline and ampicillin was lower 61.4%, 55.2% [17,18]. This perhaps means Sulfamethoxazole trimethoprim, tetracycline, ampicillin and amoxicillin need to be reviewed in the treatment of diarrhea in this population. DEC resistance was significantly higher among children below five years compared to domestic food animals that reported less than 20% resistant to only four antibiotics. This finding could be explained by the fact that gut inflammation like inflammatory diarrhea can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae [19]. In Kenya, antibiotic therapy is recommended for treatment of dysentery and salmonellosis for children below five years, while oral rehydration solutions and zinc sulphate are recommended for the management of DEC-related diarrhea (MOH 2016). However, low diagnostic capacities in lowmiddle income countries promote empiric treatment of childhood diarrhoea with antibiotics, which likely contributes to the emergence of resistant strains [20]. This study reports on low prevalence of resistance towards Gentamicin and ciprofloxacin to EAEC, EPEC and ETEC isolates which contrast certain studies in Burkhina faso [21]. The environment foremost likely permits comprehensive disease progression that lead to the clinical manifestation of resistant genes. Numerous reports advocate the global all-inclusive One Health approach to ameliorate the emergence and dissemination of antimicrobial resistance [22]. Efforts towards antibiotic management are best complemented with water, sanitation and hygiene interventions to reduce childhood diarrhea-related deaths.
This study has reported sensitivity to Merepenem 84.6%, Gentamicin 80.8%, Nalidixic acid 73.1%, Chloramphenicol 69.2%, Ceftriaxone 65.4 and Ciprofloxacin 61.5%. The findings towards merepenem sensitivity may be due to limited access as it is not prescribed at the outpatient clinics in Kenya. The susceptibility findings contradict studies conducted in India that reported high prevalence of antibiotics resistant of E.coli among a community in the rural settings, the high prevalence was mostly detected among the children [23]. This findings highlights the knowledge of the antibiogram profiles of pathogens could help understand the treatment of choice [18].
Analysis of food animal samples showed sensitivity towards most of the antibiotic that were used for treatment of DEC. This reveals the low usage of antibiotics in the food animals feeds and water in the farms by farmers for the purpose of growth promotion that creates favourable conditions for selection and spread of resistant bacteria within animals. These findings contrasts those from a study performed in south Africa that reported resistance towards similar antibiotics to food animals. However, reported resistance below 20% to Ampicillin, Azithromycin and Amoxicillin, the low resistance cannot not be ignored since it reveals that animal products pose a public health risk [11].
The growing Multidrug-Resistance (MDR) have been witnessed and broadly spreading amongst gram-negative bacteria [24]. This study reports the MDR rates of DEC isolates (70%) Sulfamethoxazole/Trimethoprim, ampicilin, Amoxicillin and tetracycline were the greatest contributors of MDR; SXT 29(100 %), Amp 28(92.3%) and tet 29(100 %) while Ceftriaxone and Nalidixic Acid contributed only 10% to MDR respectively.This means that diarrhea case management among children below five years perhaps remains with a narrow choice of treatment. This finding concurs to studies conducted in Qatar and Iran (31). However, the present finding of Multidrug-Resistant (MDR) phenotypes was lower than those reported by [27]. The current multidrug resistance is probably due to over-the-counter antibiotics that are widely available and used inappropriately by humans and on animal production farms and this bacteria are known to accumulate genes responsible for coding antimicrobial resistance mechanisms [28]. Most MDR index reported greater than 0.2 suggesting high risk of pathogenic transmission in the environment [29]. The high percentage of MDR isolates revealed by this study among the children samples suggests that these organisms were exposed to high antibiotic use in the environment. Hence, rise in MDR index exacerbate the diarrhea case management resulting to limited treatment options.
Environment contaminated with animals infected dug contaminates animals teats and udder and this is passed to the raw milk hence facilitates transmission of antimicrobial resistant bacteria to children. Additionally,the variable of ‘’children eating soil’’ during crawling as part of development exposes them to increased AMR load in the environment. Lack of personal hygienic practices such as washing of hands after use of the public toilets is associated with increased ampicillin resistant in children and increased tetracycline resistant E.coli in adults. Lastly, Sulfa drugs used as antibiotics are used as malaria treatment options in endemic areas, thus this practice correlates to load of Sulfamethoxazole resistant E.coli in children and it is revealed as a contributing factor to resistant [30].
Our study had few limitations. Isolates analysed from this study were collected from sick children who visited the health facility and they represent a non-randomized subset of the population. Consequently, our results may not generalize the entire children population. Lastly, given that DEC strains isolates are secondary cause of diarrhea among children we were not able to conclusively assess the severity of the pathogens in the population. Hence, longitudinal cohort studies that include a randomized population of children may be suited to address the limitation.
Conclusion
High prevalence of multidrug resistance was reported among children compared to food animals suggesting possibility of transmission of multidrug resistance bacteria between children and food animals. Therefore, Global and National Action plan need to re-direct its effort to reduce the burden of antimicrobial resistance in-order to attain optimal health in children, animals and environment.
Declarations
Ethical approval: This study was approved by Jaramogi Oginga Odinga Ethical Review Board REF: ISERC/JOOTRH/600/22. Confidentiality was observed throughout the study period.
Competing interest: Authors have no competing interest.
Consent to participate: Informed consent was obtained from all participants involved in the study.
Authors’ contributions: R.Y Conceptualization and writing – original draft, R.Y, E.K & C.O Methodology, data curation formal analysis. G.M, J.G & G.K Supervision and writing-review and editing.
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399(10325): 629-655.
- Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in foodproducing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health. 2017; 1(8): 316-327.
- Woolhouse M, Ward M, van Bunnik B, Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc Lond B Biol Sci. 2015; 370(1670): 20140083.
- Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog Glob Health. 2015; 109(7): 309-318.
- Khairy RMM, Fathy ZA, Mahrous DM, Mohamed ES, Abdelrahim SS. Prevalence, phylogeny, and antimicrobial resistance of Escherichia coli pathotypes isolated from children less than 5 years old with community acquired- diarrhea in Upper Egypt. BMC Infectious Diseases. 2020; 20(1): 908.
- Salleh MZ, Nik Zuraina NMN, Hajissa K, Ilias MI, Deris ZZ. Prevalence of Multidrug-Resistant Diarrheagenic Escherichia coli in Asia: A Systematic Review and Meta-Analysis. Antibiotics (Basel). 2022; 11(10): 1333.
- Collignon P, Beggs JJ, Walsh TR, Gandra S, Laxminarayan R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet Health. 2018; 2(9): 398-405.
- Baker KK, Mumma JAO, Simiyu S, Sewell D, Tsai K, Anderson JD, et al. Environmental and behavioural exposure pathways associated with diarrhoea and enteric pathogen detection in 5-month-old, periurban Kenyan infants: a cross-sectional study. BMJ Open. 2022; 12(10): 059878.
- Kk B, Jao M, S S, D S, K T, Jd A, et al. Environmental and behavioural exposure pathways associated with diarrhoea and enteric pathogen detection in 5-month-old, periurban Kenyan infants: a cross-sectional study. BMJ open. 2022; 12(10). doi:10.1136/bmjopen-2021-059878.
- Kipkirui E, Koech M, Ombogo A, Kirera R, Ndonye J, Kipkemoi N, et al. Molecular characterization of enterotoxigenic Escherichia coli toxins and colonization factors in children under five years with acute diarrhea attending Kisii Teaching and Referral Hospital, Kenya. Trop Dis Travel Med Vaccines. 2021; 7(1): 31.
- Abdalla SE, Abia ALK, Amoako DG, Perrett K, Bester LA, Essack SY. From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use. Antibiotics (Basel). 2021; 10(2): 178.
- Wolde A, Deneke Y, Sisay T, Mathewos M. Molecular Characterization and Antimicrobial Resistance of Pathogenic Escherichia coli Strains in Children from Wolaita Sodo, Southern Ethiopia. J Trop Med. 2022; 2022: 9166209.
- Zelelie TZ, Eguale T, Yitayew B, Abeje D, Alemu A, Seman A, et al. Molecular epidemiology and antimicrobial susceptibility of diarrheagenic Escherichia coli isolated from children under age five with and without diarrhea in Central Ethiopia. PLoS One. 2023; 18(7): 0288517.
- Saka HK, Dabo NT, Muhammad B, García-Soto S, Ugarte-Ruiz M, Alvarez J. Diarrheagenic Escherichia coli Pathotypes From Children Younger Than 5 Years in Kano State, Nigeria. Front Public Health. 2019; 7: 348.
- Verma S, Venkatesh V, Kumar R, Kashyap S, Kumar M, Maurya AK, et al. Etiological agents of diarrhea in hospitalized pediatric patients with special emphasis on diarrheagenic Escherichia coli in North India. J Lab Physicians. 2019; 11(1): 68-74.
- Sang W, Too R, Githii S, Githui W, Wanzala P, Kariuki J, et al. Emerging Antimicrobial Resistance Patterns of Enteric Pathogens Isolated from Children under 5 years in EAPHLNP Satellite Sites in Kenya. African Journal of Health Sciences. 2019; 32(6): 47-58.
- Nkatha ML, Kangogo M, Waititu KK, Sang WK. Characterization of Selected Escherichia coli Pathovars and Their Antimicrobial Resistance Patterns among Diarrheal Children under the Age of Five Years from Machakos County, Kenya. American Journal of Infectious Diseases and Microbiology. 2021; 9(2): 51-55.
- Belete MA, Demlie TB, Chekole WS, Sisay Tessema T. Molecular identification of diarrheagenic Escherichia coli pathotypes and their antibiotic resistance patterns among diarrheic children and in contact calves in Bahir Dar city, Northwest Ethiopia. PLoS One. 2022; 17(9): 0275229.
- Kariuki S, Mbae C, Onsare R, Kavai SM, Wairimu C, Ngetich R, et al. Multidrug-resistant Nontyphoidal Salmonella Hotspots as Targets for Vaccine Use in Management of Infections in Endemic Settings. Clin Infect Dis. 2019; 68(l1): 10-15.
- Kariuki S, Wairimu C, Mbae C. Antimicrobial Resistance in Endemic Enteric Infections in Kenya and the Region, and Efforts Toward Addressing the Challenges. J Infect Dis. 2021; 224(7): 883-889.
- Konaté A, Dembélé R, Guessennd NK, Kouadio FK, Kouadio IK, Ouattara MB, et al. Epidemiology and Antibiotic Resistance Phenotypes of Diarrheagenic Escherichia coli Responsible for Infantile Gastroenteritis in Ouagadougou, Burkina Faso. Eur J Microbiol Immunol (Bp). 2017; 7(3): 168-175.
- Thakur S, Gray GC. The Mandate for a Global ‘One Health’ Approach to Antimicrobial Resistance Surveillance. Am J Trop Med Hyg. 2019; 100(2): 227-228.
- Purohit MR, Lindahl LF, Diwan V, Marrone G, Lundborg CS. High levels of drug resistance in commensal E. coli in a cohort of children from rural central India. Sci Rep. 2019; 9(1): 6682.
- Eltai NO, Al Thani AA, Al Hadidi SH, Al Ansari K, Yassine HM. Antibiotic resistance and virulence patterns of pathogenic Escherichia coli strains associated with acute gastroenteritis among children in Qatar. BMC Microbiol. 2020; 20(1): 54.
- Mahdavi Broujerdi S, Roayaei Ardakani M, Rezatofighi SE. Characterization of diarrheagenic Escherichia coli strains associated with diarrhea in children, Khouzestan, Iran. J Infect Dev Ctries. 2018; 12(8): 649-656.
- Srivani M, Reddy YN, Subramanyam KV, Reddy MR, Rao TS. Prevalence and antimicrobial resistance pattern of Shiga toxigenic Escherichia coli in diarrheic buffalo calves. Vet World. 2017; 10(7): 774-778.
- Yang G-Y, Guo L, Su J-H, Zhu Y-H, Jiao L-G, Wang J-F. Frequency of Diarrheagenic Virulence Genes and Characteristics in Escherichia coli Isolates from Pigs with Diarrhea in China. Microorganisms. 2019; 7(9): 308.
- Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules. 2018; 23(4): 795.
- Teshome A, Alemayehu T, Deriba W, Ayele Y. Antibiotic Resistance Profile of Bacteria Isolated from Wastewater Systems in Eastern Ethiopia. J Environ Public Health. 2020; 2020: 2796365.
- Omulo S, Lofgren ET, Lockwood S, Thumbi SM, Bigogo G, Ouma A, et al. Carriage of antimicrobial-resistant bacteria in a highdensity informal settlement in Kenya is associated with environmental risk-factors. Antimicrob Resist Infect Control. 2021; 10(1): 18.